7cd05162e9bc9af26f65cc72bfa23a4eb08c79f8,skbio/draw/tests/test_distributions.py,DistributionsTests,test_get_distribution_markers_insufficient_markers,#DistributionsTests#,133
Before Change
["b", "g", "r", "c"])
def test_get_distribution_markers_insufficient_markers(self):
self.assertEqual(npt.assert_warns(RuntimeWarning,
_get_distribution_markers,
"colors", None, 10),
["b", "g", "r", "c", "m", "y", "w", "b", "g", "r"])
self.assertEqual(npt.assert_warns(RuntimeWarning,
_get_distribution_markers,
"symbols", ["^", ">", "<"], 5),
["^", ">", "<", "^", ">"])
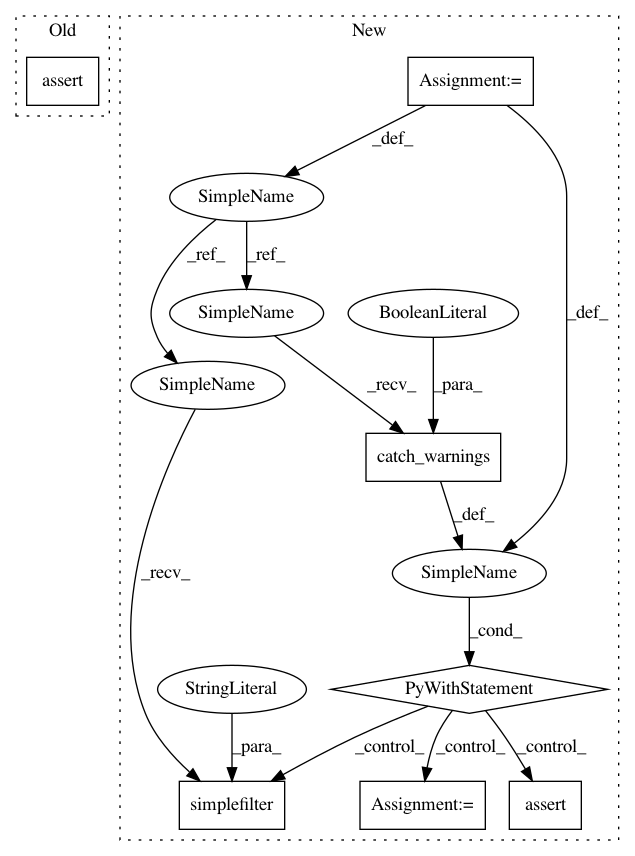
After Change
expected = ["b", "g", "r", "c", "m", "y", "w", "b", "g", "r"]
// adapted from SO example here: http://stackoverflow.com/a/3892301
with warnings.catch_warnings(record=True) as w:
warnings.simplefilter("always")
actual = _get_distribution_markers("colors", None, 10)
self.assertTrue(issubclass(w[-1].category, RuntimeWarning))
self.assertEqual(actual, expected)
expected = ["^", ">", "<", "^", ">"]
with warnings.catch_warnings(record=True) as w:
warnings.simplefilter("always")
actual = _get_distribution_markers("symbols", ["^", ">", "<"], 5)
In pattern: SUPERPATTERN
Frequency: 4
Non-data size: 7
Instances
Project Name: biocore/scikit-bio
Commit Name: 7cd05162e9bc9af26f65cc72bfa23a4eb08c79f8
Time: 2015-06-26
Author: gregcaporaso@gmail.com
File Name: skbio/draw/tests/test_distributions.py
Class Name: DistributionsTests
Method Name: test_get_distribution_markers_insufficient_markers
Project Name: biocore/scikit-bio
Commit Name: 7cd05162e9bc9af26f65cc72bfa23a4eb08c79f8
Time: 2015-06-26
Author: gregcaporaso@gmail.com
File Name: skbio/draw/tests/test_distributions.py
Class Name: DistributionsTests
Method Name: test_get_distribution_markers_insufficient_markers
Project Name: biocore/scikit-bio
Commit Name: d7e3e14729b53ef09d8ad32cc6925c285d42eabc
Time: 2015-06-26
Author: gregcaporaso@gmail.com
File Name: skbio/draw/tests/test_distributions.py
Class Name: DistributionsTests
Method Name: test_grouped_distributions_insufficient_symbols
Project Name: biocore/scikit-bio
Commit Name: 7cd05162e9bc9af26f65cc72bfa23a4eb08c79f8
Time: 2015-06-26
Author: gregcaporaso@gmail.com
File Name: skbio/draw/tests/test_distributions.py
Class Name: DistributionsTests
Method Name: test_grouped_distributions_insufficient_colors
Project Name: biocore/scikit-bio
Commit Name: d7e3e14729b53ef09d8ad32cc6925c285d42eabc
Time: 2015-06-26
Author: gregcaporaso@gmail.com
File Name: skbio/draw/tests/test_distributions.py
Class Name: DistributionsTests
Method Name: test_set_figure_size_long_labels
