4b1f799c8403626ff56c7bf23fb1fbdb79dcb015,scanpy/plotting/_tools/__init__.py,,_rank_genes_groups_plot,#Any#Any#Any#Any#Any#Any#Any#Any#,258
Before Change
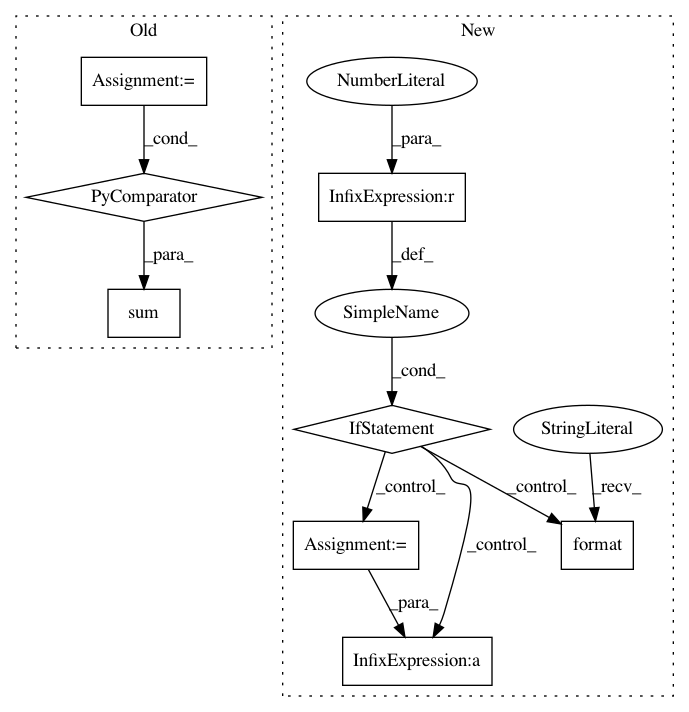
group_positions = [(x, x + n_genes - 1) for x in range(0, n_genes * len(group_names), n_genes)]
// sum(list, []) is used to flatten the gene list
gene_names = sum([list(adata.uns[key]["names"][x][:n_genes]) for x in group_names], [])
if plot_type == "dotplot":
from .._anndata import dotplot
dotplot(adata, gene_names, groupby, var_group_labels=group_names,After Change
for group in group_names:
// get all genes that are "not-nan"
genes_list = [gene for gene in adata.uns[key]["names"][group] if not pd.isnull(gene)][:n_genes]
if len(genes_list) == 0:
logg.warn("No genes found for group {}".format(group))
continue
gene_names.extend(genes_list)
end = start + len(genes_list)
group_positions.append((start, end -1 ))
group_names_valid.append(group)
start = end
In pattern: SUPERPATTERN
Frequency: 3
Non-data size: 8
Instances Project Name: theislab/scanpy
Commit Name: 4b1f799c8403626ff56c7bf23fb1fbdb79dcb015
Time: 2019-01-14
Author: fidel.ramirez@gmail.com
File Name: scanpy/plotting/_tools/__init__.py
Class Name:
Method Name: _rank_genes_groups_plot
Project Name: KaiyangZhou/deep-person-reid
Commit Name: b88d36cd9c8056e15607a40f5d10a9072ab84b22
Time: 2018-07-06
Author: k.zhou@qmul.ac.uk
File Name: train_imgreid_xent.py
Class Name:
Method Name: main
Project Name: KaiyangZhou/deep-person-reid
Commit Name: b88d36cd9c8056e15607a40f5d10a9072ab84b22
Time: 2018-07-06
Author: k.zhou@qmul.ac.uk
File Name: train_vidreid_xent.py
Class Name:
Method Name: main