77550cc1f3ccf9537265d127b0f25aa790d2b4b5,skopt/acquisition.py,,gaussian_pi,#Any#Any#Any#Any#Any#,149
Before Change
mu, std = model.predict(X, return_std=True)
// make sure mu and std have the same shape so we can divide them below
std = std.reshape(mu.shape)
values = np.zeros_like(mu)
mask = std > 0
improve = y_opt - xi - mu[mask]After Change
mu, std = model.predict(X, return_std=True)
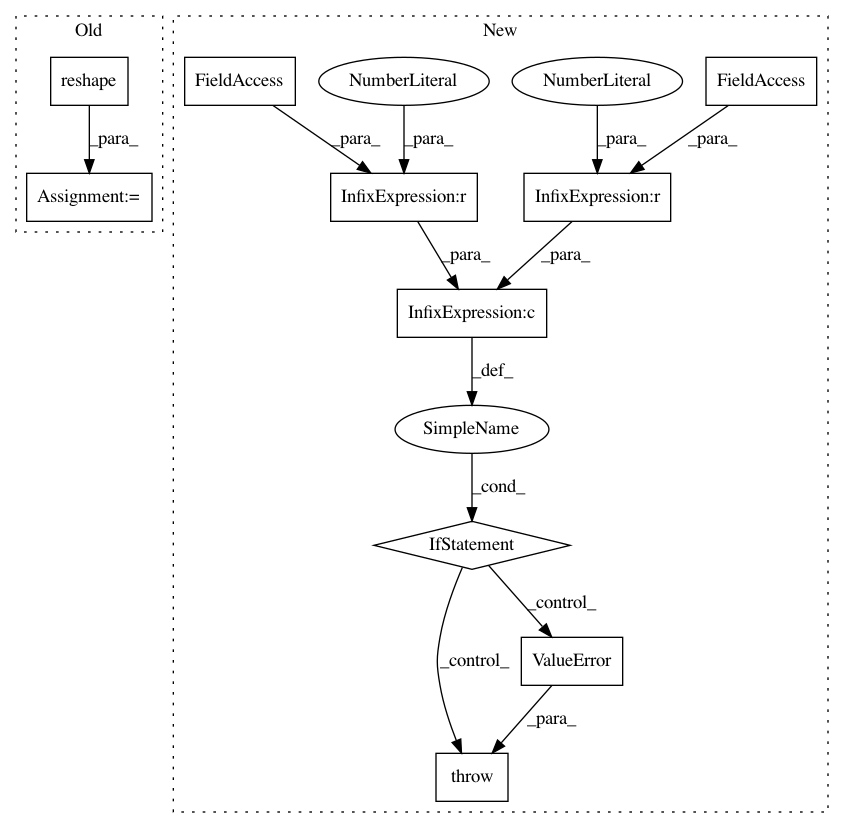
// check dimensionality of mu, std so we can divide them below
if (mu.ndim != 1) or (std.ndim != 1):
raise ValueError("mu and std are {}-dimensional and {}-dimensional, "
"however both must be 1-dimensional. Did you train "
"your model with an (N, 1) vector instead of an "
"(N,) vector?"
.format(mu.ndim, std.ndim))
values = np.zeros_like(mu)
mask = std > 0
improve = y_opt - xi - mu[mask]
scaled = improve / std[mask]In pattern: SUPERPATTERN
Frequency: 3
Non-data size: 10
Instances Project Name: scikit-optimize/scikit-optimize
Commit Name: 77550cc1f3ccf9537265d127b0f25aa790d2b4b5
Time: 2018-11-07
Author: hinnefe2@illinois.edu
File Name: skopt/acquisition.py
Class Name:
Method Name: gaussian_pi
Project Name: scikit-optimize/scikit-optimize
Commit Name: 77550cc1f3ccf9537265d127b0f25aa790d2b4b5
Time: 2018-11-07
Author: hinnefe2@illinois.edu
File Name: skopt/acquisition.py
Class Name:
Method Name: gaussian_ei
Project Name: pymc-devs/pymc3
Commit Name: 6d0496672093058ef2a40c1123c3cd824df31471
Time: 2017-06-09
Author: adrian.seyboldt@gmail.com
File Name: pymc3/distributions/multivariate.py
Class Name: MvNormal
Method Name: logp