53059cec9053a8429b7f0a8f06e6cbe1672b60e7,cellprofiler/modules/tests/test_groups.py,TestGroups,test_02_01_group_on_two,#TestGroups#,144
Before Change
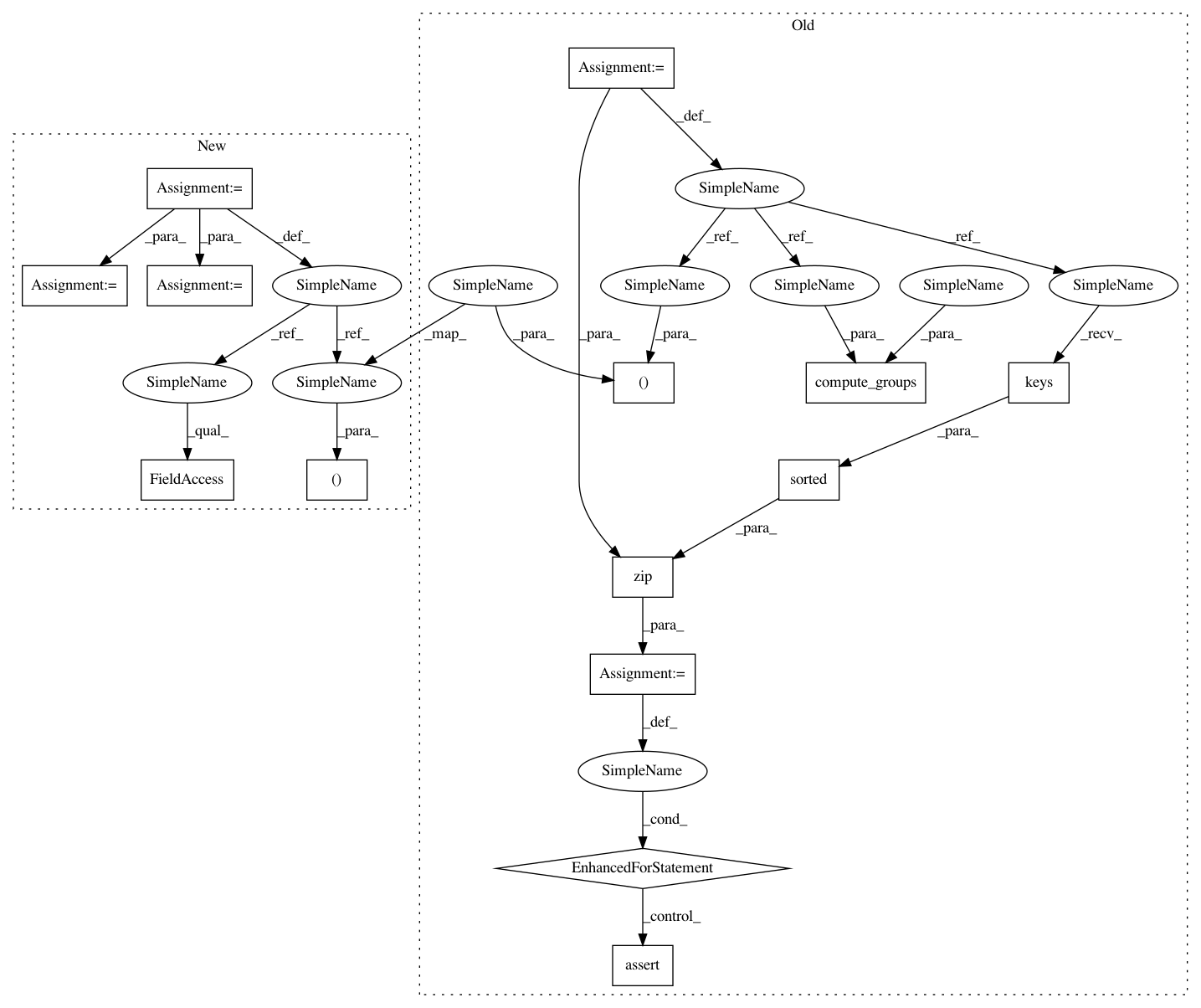
groups.add_grouping_metadata()
groups.grouping_metadata[1].image_name.value = "DNA"
groups.grouping_metadata[1].metadata_choice.value = "Site"
iscds, image_set_key_names, image_sets = self.make_image_sets(
(("Plate", ("P-12345", "P-23456")),
("Well", ("A01", "A02", "A03")),
("Site", ("1", "2", "3", "4"))),
(("DNA", "Wavelength", "1", cpp.Pipeline.ImageSetChannelDescriptor.CT_GRAYSCALE),
("GFP", "Wavelength", "1", cpp.Pipeline.ImageSetChannelDescriptor.CT_GRAYSCALE)))
key_list, groupings = groups.compute_groups(
iscds, image_set_key_names, image_sets)
self.assertEqual(len(key_list), 2)
self.assertEqual(key_list[0], (1, "Plate"))
self.assertEqual(key_list[1], (0, "Site"))
self.assertEqual(len(groupings), 2 * 4)
for plate in ("P-12345", "P-23456"):
for site in ("1", "2", "3", "4"):
self.assertTrue(groupings.has_key((plate, site)))
image_set_list = groupings[(plate, site)]
self.assertEqual(len(image_set_list), 3)
expected_image_set_list = \
sorted(filter(lambda x: x[0] == plate and x[2] == site,
image_sets.keys()))
for image_set_key, expected_image_set_key in zip(
image_set_list, expected_image_set_list):
self.assertEqual(image_set_key, expected_image_set_key)
After Change
m[cpmeas.IMAGE, ftr, image_number].startswith(plate))
def test_02_01_group_on_two(self):
groups, workspace = self.make_image_sets(
(("Plate", ("P-12345", "P-23456")),
("Well", ("A01", "A02", "A03")),
("Site", ("1", "2", "3", "4"))),
(("DNA", "Wavelength", "1", cpp.Pipeline.ImageSetChannelDescriptor.CT_GRAYSCALE),
("GFP", "Wavelength", "1", cpp.Pipeline.ImageSetChannelDescriptor.CT_GRAYSCALE)))
groups.wants_groups.value = True
groups.grouping_metadata[0].metadata_choice.value = "Plate"
groups.add_grouping_metadata()
groups.grouping_metadata[1].metadata_choice.value = "Site"
self.assertTrue(groups.prepare_run(workspace))
m = workspace.measurements
assert isinstance(m, cpmeas.Measurements)
image_numbers = m.get_image_numbers()
pipeline = workspace.pipeline
assert isinstance(pipeline, cpp.Pipeline)
key_list, groupings = pipeline.get_groupings(workspace)
self.assertEqual(len(key_list), 2)
self.assertEqual(key_list[0], "Metadata_Plate")
self.assertEqual(key_list[1], "Metadata_Site")
self.assertEqual(len(groupings), 8)
idx = 0;
for plate in ("P-12345", "P-23456"):
for site in ("1", "2", "3", "4"):
grouping, image_set_list = groupings[idx]
idx += 1
self.assertEqual(grouping["Metadata_Plate"], plate)
self.assertEqual(grouping["Metadata_Site"], site)
self.assertEqual(len(image_set_list), 3)
ftr = "_".join((cpmeas.C_FILE_NAME, "DNA"))
for image_number in image_set_list:
file_name = m[cpmeas.IMAGE, ftr, image_number]
p, w, s, rest = file_name.split("_")
self.assertEqual(p, plate)
self.assertEqual(s, site)
In pattern: SUPERPATTERN
Frequency: 3
Non-data size: 14
Instances
Project Name: CellProfiler/CellProfiler
Commit Name: 53059cec9053a8429b7f0a8f06e6cbe1672b60e7
Time: 2013-01-16
Author: leek@broadinstitute.org
File Name: cellprofiler/modules/tests/test_groups.py
Class Name: TestGroups
Method Name: test_02_01_group_on_two
Project Name: CellProfiler/CellProfiler
Commit Name: 53059cec9053a8429b7f0a8f06e6cbe1672b60e7
Time: 2013-01-16
Author: leek@broadinstitute.org
File Name: cellprofiler/modules/tests/test_groups.py
Class Name: TestGroups
Method Name: test_02_01_group_on_two
Project Name: CellProfiler/CellProfiler
Commit Name: 53059cec9053a8429b7f0a8f06e6cbe1672b60e7
Time: 2013-01-16
Author: leek@broadinstitute.org
File Name: cellprofiler/modules/tests/test_groups.py
Class Name: TestGroups
Method Name: test_02_00_compute_no_groups
Project Name: CellProfiler/CellProfiler
Commit Name: 53059cec9053a8429b7f0a8f06e6cbe1672b60e7
Time: 2013-01-16
Author: leek@broadinstitute.org
File Name: cellprofiler/modules/tests/test_groups.py
Class Name: TestGroups
Method Name: test_02_01_group_on_one
