c6f415ebddd6f765e1d5e508aaddba0c38fa392a,skbio/core/sequence.py,NucleotideSequence,iupac_degeneracies,#Any#,1015
Before Change
non-degenerate IUPAC nucleotide characters it represents.
return {
"R": set("AG"), "Y": set("CTU"), "M": set("AC"), "K": set("TUG"),
"W": set("ATU"), "S": set("GC"), "B": set("CGTU"),
"D": set("AGTU"), "H": set("ACTU"), "V": set("ACG"),
"N": set("ACGTU")
}
@classmethod
def iupac_degenerate_characters(cls):
Return the degenerate IUPAC nucleotide characters.
After Change
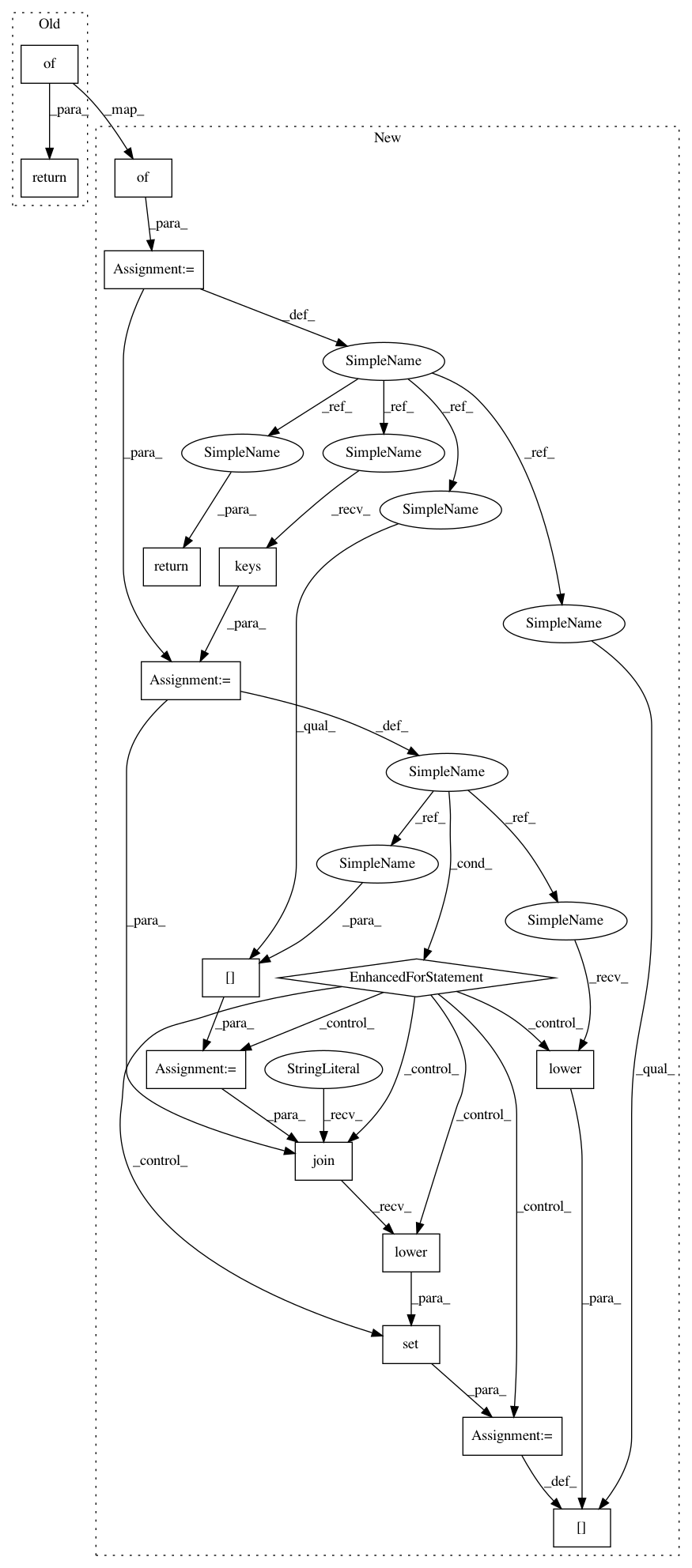
non-degenerate IUPAC nucleotide characters it represents.
degen_map = {
"R": set("AG"), "Y": set("CTU"), "M": set("AC"), "K": set("TUG"),
"W": set("ATU"), "S": set("GC"), "B": set("CGTU"),
"D": set("AGTU"), "H": set("ACTU"), "V": set("ACG"),
"N": set("ACGTU")
}
for degen_char in degen_map.keys():
nondegen_chars = degen_map[degen_char]
degen_map[degen_char.lower()] = set(
"".join(nondegen_chars).lower())
return degen_map
@classmethod
def iupac_degenerate_characters(cls):
Return the degenerate IUPAC nucleotide characters.
In pattern: SUPERPATTERN
Frequency: 3
Non-data size: 16
Instances
Project Name: biocore/scikit-bio
Commit Name: c6f415ebddd6f765e1d5e508aaddba0c38fa392a
Time: 2014-03-21
Author: jai.rideout@gmail.com
File Name: skbio/core/sequence.py
Class Name: NucleotideSequence
Method Name: iupac_degeneracies
Project Name: biocore/scikit-bio
Commit Name: c6f415ebddd6f765e1d5e508aaddba0c38fa392a
Time: 2014-03-21
Author: jai.rideout@gmail.com
File Name: skbio/core/sequence.py
Class Name: RNASequence
Method Name: iupac_degeneracies
Project Name: biocore/scikit-bio
Commit Name: c6f415ebddd6f765e1d5e508aaddba0c38fa392a
Time: 2014-03-21
Author: jai.rideout@gmail.com
File Name: skbio/core/sequence.py
Class Name: DNASequence
Method Name: iupac_degeneracies
Project Name: biocore/scikit-bio
Commit Name: c6f415ebddd6f765e1d5e508aaddba0c38fa392a
Time: 2014-03-21
Author: jai.rideout@gmail.com
File Name: skbio/core/sequence.py
Class Name: NucleotideSequence
Method Name: iupac_degeneracies
