da6fb45ee7a00f5051ec47bf15247045774c3604,mimic3benchmark/scripts/create_decompensation.py,,process_partition,#Any#Any#Any#Any#Any#Any#,13
Before Change
xty_triples = []
patients = list(filter(str.isdigit, os.listdir(os.path.join(args.root_path, partition))))
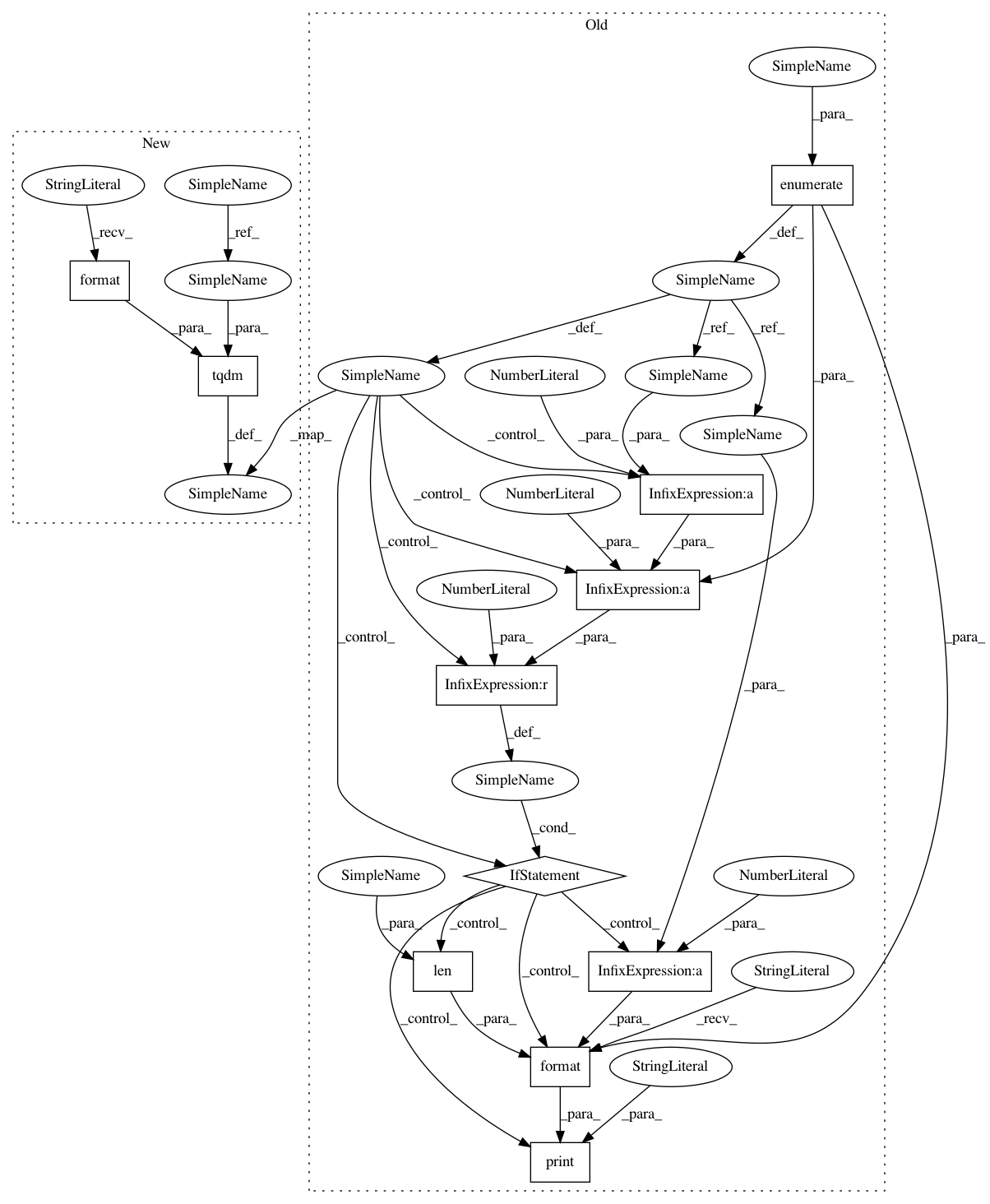
for (patient_index, patient) in enumerate(patients):
patient_folder = os.path.join(args.root_path, partition, patient)
patient_ts_files = list(filter(lambda x: x.find("timeseries") != -1, os.listdir(patient_folder)))
stays_df = pd.read_csv(os.path.join(patient_folder, "stays.csv"))
for ts_filename in patient_ts_files:
with open(os.path.join(patient_folder, ts_filename)) as tsfile:
lb_filename = ts_filename.replace("_timeseries", "")
label_df = pd.read_csv(os.path.join(patient_folder, lb_filename))
// empty label file
if label_df.shape[0] == 0:
continue
mortality = int(label_df.iloc[0]["Mortality"])
los = 24.0 * label_df.iloc[0]["Length of Stay"] // in hours
if pd.isnull(los):
print("(length of stay is missing)", patient, ts_filename)
continue
stay = stays_df[stays_df.ICUSTAY_ID == label_df.iloc[0]["Icustay"]]
deathtime = stay["DEATHTIME"].iloc[0]
intime = stay["INTIME"].iloc[0]
if pd.isnull(deathtime):
lived_time = 1e18
else:
lived_time = (datetime.strptime(deathtime, "%Y-%m-%d %H:%M:%S") -
datetime.strptime(intime, "%Y-%m-%d %H:%M:%S")).total_seconds() / 3600.0
ts_lines = tsfile.readlines()
header = ts_lines[0]
ts_lines = ts_lines[1:]
event_times = [float(line.split(",")[0]) for line in ts_lines]
ts_lines = [line for (line, t) in zip(ts_lines, event_times)
if -eps < t < los + eps]
event_times = [t for t in event_times
if -eps < t < los + eps]
// no measurements in ICU
if len(ts_lines) == 0:
print("(no events in ICU) ", patient, ts_filename)
continue
sample_times = np.arange(0.0, min(los, lived_time) + eps, sample_rate)
sample_times = list(filter(lambda x: x > shortest_length, sample_times))
// At least one measurement
sample_times = list(filter(lambda x: x > event_times[0], sample_times))
output_ts_filename = patient + "_" + ts_filename
with open(os.path.join(output_dir, output_ts_filename), "w") as outfile:
outfile.write(header)
for line in ts_lines:
outfile.write(line)
for t in sample_times:
if mortality == 0:
cur_mortality = 0
else:
cur_mortality = int(lived_time - t < future_time_interval)
xty_triples.append((output_ts_filename, t, cur_mortality))
if (patient_index + 1) % 100 == 0:
print("processed {} / {} patients".format(patient_index + 1, len(patients)), end="\r")
print(len(xty_triples))
if partition == "train":
random.shuffle(xty_triples)
if partition == "test":
After Change
os.mkdir(output_dir)
xty_triples = []
patients = list(filter(str.isdigit, os.listdir(os.path.join(args.root_path, partition))))
for patient in tqdm(patients, desc="Iterating over patients in {}".format(partition)):
patient_folder = os.path.join(args.root_path, partition, patient)
patient_ts_files = list(filter(lambda x: x.find("timeseries") != -1, os.listdir(patient_folder)))
stays_df = pd.read_csv(os.path.join(patient_folder, "stays.csv"))
In pattern: SUPERPATTERN
Frequency: 4
Non-data size: 11
Instances
Project Name: YerevaNN/mimic3-benchmarks
Commit Name: da6fb45ee7a00f5051ec47bf15247045774c3604
Time: 2020-03-26
Author: harhro@gmail.com
File Name: mimic3benchmark/scripts/create_decompensation.py
Class Name:
Method Name: process_partition
Project Name: YerevaNN/mimic3-benchmarks
Commit Name: da6fb45ee7a00f5051ec47bf15247045774c3604
Time: 2020-03-26
Author: harhro@gmail.com
File Name: mimic3benchmark/scripts/create_length_of_stay.py
Class Name:
Method Name: process_partition
Project Name: YerevaNN/mimic3-benchmarks
Commit Name: da6fb45ee7a00f5051ec47bf15247045774c3604
Time: 2020-03-26
Author: harhro@gmail.com
File Name: mimic3benchmark/scripts/create_in_hospital_mortality.py
Class Name:
Method Name: process_partition
Project Name: YerevaNN/mimic3-benchmarks
Commit Name: da6fb45ee7a00f5051ec47bf15247045774c3604
Time: 2020-03-26
Author: harhro@gmail.com
File Name: mimic3benchmark/scripts/create_phenotyping.py
Class Name:
Method Name: process_partition
