ebd36d3cad0d6daa604e93ed43a7a0c3b7250baf,skbio/io/fastq.py,,_fastq_to_nucleotide_sequence,#Any#Any#Any#Any#,340
Before Change
@register_reader("fastq", NucleotideSequence)
def _fastq_to_nucleotide_sequence(fh, variant=None, phred_offset=None,
seq_num=1):
return _fastq_to_sequence(fh, variant, phred_offset, seq_num,
NucleotideSequence)
@register_reader("fastq", DNASequence)
def _fastq_to_dna_sequence(fh, variant=None, phred_offset=None, seq_num=1):
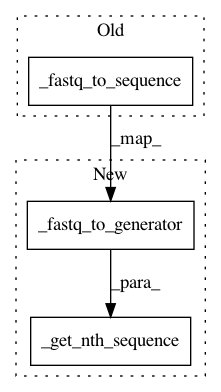
After Change
@register_reader("fastq", NucleotideSequence)
def _fastq_to_nucleotide_sequence(fh, variant=None, phred_offset=None,
seq_num=1):
return _get_nth_sequence(
_fastq_to_generator(fh, variant=variant, phred_offset=phred_offset,
constructor=NucleotideSequence),
seq_num)
@register_reader("fastq", DNASequence)
def _fastq_to_dna_sequence(fh, variant=None, phred_offset=None, seq_num=1):
In pattern: SUPERPATTERN
Frequency: 5
Non-data size: 3
Instances
Project Name: biocore/scikit-bio
Commit Name: ebd36d3cad0d6daa604e93ed43a7a0c3b7250baf
Time: 2014-10-25
Author: jai.rideout@gmail.com
File Name: skbio/io/fastq.py
Class Name:
Method Name: _fastq_to_nucleotide_sequence
Project Name: biocore/scikit-bio
Commit Name: ebd36d3cad0d6daa604e93ed43a7a0c3b7250baf
Time: 2014-10-25
Author: jai.rideout@gmail.com
File Name: skbio/io/fastq.py
Class Name:
Method Name: _fastq_to_biological_sequence
Project Name: biocore/scikit-bio
Commit Name: ebd36d3cad0d6daa604e93ed43a7a0c3b7250baf
Time: 2014-10-25
Author: jai.rideout@gmail.com
File Name: skbio/io/fastq.py
Class Name:
Method Name: _fastq_to_rna_sequence
Project Name: biocore/scikit-bio
Commit Name: ebd36d3cad0d6daa604e93ed43a7a0c3b7250baf
Time: 2014-10-25
Author: jai.rideout@gmail.com
File Name: skbio/io/fastq.py
Class Name:
Method Name: _fastq_to_dna_sequence
Project Name: biocore/scikit-bio
Commit Name: ebd36d3cad0d6daa604e93ed43a7a0c3b7250baf
Time: 2014-10-25
Author: jai.rideout@gmail.com
File Name: skbio/io/fastq.py
Class Name:
Method Name: _fastq_to_protein_sequence
