9fb388e18273e003a37e71f1dfcc663094bba754,PyMC2/sandbox/GaussianProcess/Covariance.py,Covariance,__call__,#Covariance#Any#Any#,86
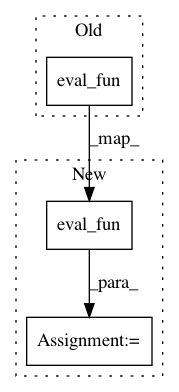
Before Change
if y is None:
C = asmatrix(zeros((lenx,lenx)),dtype=float)
self.eval_fun(C,x,x,**self.params)
if not self.conditioned:
return C
RF = asmatrix(zeros((lenx,self.obs_len),dtype=float))After Change
if y is None:
C=self.eval_fun(x,x,**self.params)
if not self.conditioned:
return C
RF = self.eval_fun(x, self.obs_mesh, **self.params) In pattern: SUPERPATTERN
Frequency: 3
Non-data size: 3
Instances Project Name: pymc-devs/pymc3
Commit Name: 9fb388e18273e003a37e71f1dfcc663094bba754
Time: 2007-04-04
Author: anand.prabhakar.patil@15d7aa0b-6f1a-0410-991a-d59f85d14984
File Name: PyMC2/sandbox/GaussianProcess/Covariance.py
Class Name: Covariance
Method Name: __call__
Project Name: pymc-devs/pymc3
Commit Name: 9fb388e18273e003a37e71f1dfcc663094bba754
Time: 2007-04-04
Author: anand.prabhakar.patil@15d7aa0b-6f1a-0410-991a-d59f85d14984
File Name: PyMC2/sandbox/GaussianProcess/Covariance.py
Class Name: Covariance
Method Name: __new__
Project Name: pfnet-research/chainer-chemistry
Commit Name: b38efd0d693545672eb091e8f615d92c42c7058b
Time: 2018-12-09
Author: acc1ssnn9terias@gmail.com
File Name: chainer_chemistry/saliency/calculator/integrated_gradients_calculator.py
Class Name: IntegratedGradientsCalculator
Method Name: _compute_core