f921a41292d13178a6eca168b8bd261f190b76b4,ilastik-shell/applets/dataSelection/dataSelectionSerializer.py,DataSelectionSerializer,_serializeToHdf5,#DataSelectionSerializer#Any#Any#Any#,39
Before Change
del infoDir[infoName]
// Rebuild the list of infos
for index, slot in enumerate(self.mainOperator.Dataset):
infoGroup = infoDir.create_group("info{:04d}".format(index))
datasetInfo = slot.value
locationString = self.LocationStrings[datasetInfo.location]
infoGroup.create_dataset("location", data=locationString)
infoGroup.create_dataset("filePath", data=datasetInfo.filePath)
infoGroup.create_dataset("datasetId", data=datasetInfo.datasetId)
infoGroup.create_dataset("allowLabels", data=datasetInfo.allowLabels)
// Write any missing local datasets to the local_data group
localDataGroup = self.getOrCreateGroup(topGroup, "local_data")
wroteInternalData = False
for index, slot in enumerate(self.mainOperator.Dataset):
info = slot.value
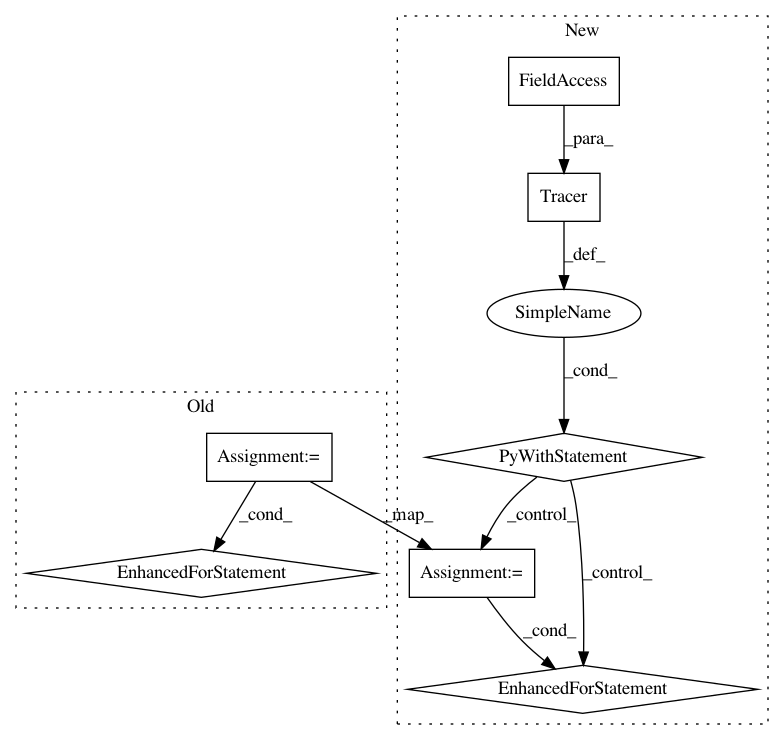
After Change
self.mainOperator.Dataset.notifyInserted( bind(handleNewDataset) )
def _serializeToHdf5(self, topGroup, hdf5File, projectFilePath):
with Tracer(traceLogger):
// If the operator has a some other project file, something"s wrong
if self.mainOperator.ProjectFile.connected():
assert self.mainOperator.ProjectFile.value == hdf5File
// Access the info group
infoDir = self.getOrCreateGroup(topGroup, "infos")
// Delete all infos
for infoName in infoDir.keys():
del infoDir[infoName]
// Rebuild the list of infos
for index, slot in enumerate(self.mainOperator.Dataset):
infoGroup = infoDir.create_group("info{:04d}".format(index))
datasetInfo = slot.value
locationString = self.LocationStrings[datasetInfo.location]
infoGroup.create_dataset("location", data=locationString)
infoGroup.create_dataset("filePath", data=datasetInfo.filePath)
infoGroup.create_dataset("datasetId", data=datasetInfo.datasetId)
infoGroup.create_dataset("allowLabels", data=datasetInfo.allowLabels)
// Write any missing local datasets to the local_data group
localDataGroup = self.getOrCreateGroup(topGroup, "local_data")
wroteInternalData = False
for index, slot in enumerate(self.mainOperator.Dataset):
info = slot.value
// If this dataset should be stored in the project, but it isn"t there yet
if info.location == DatasetInfo.Location.ProjectInternal \
and info.datasetId not in localDataGroup.keys():
// Obtain the data from the corresponding output and store it to the project.
// TODO: Optimize this for large datasets by streaming it chunk-by-chunk.
dataSlot = self.mainOperator.Image[index]
data = dataSlot[...].wait()
// Vigra thinks its okay to reorder the data if it has axistags,
// but we don"t want that. To avoid reordering, we write the data
// ourselves and attach the axistags afterwards.
dataset = localDataGroup.create_dataset(info.datasetId, data=data)
dataset.attrs["axistags"] = dataSlot.meta.axistags.toJSON()
wroteInternalData = True
// Construct a list of all the local dataset ids we want to keep
localDatasetIds = [ slot.value.datasetId
for index, slot
in enumerate(self.mainOperator.Dataset)
if slot.value.location == DatasetInfo.Location.ProjectInternal ]
// Delete any datasets in the project that aren"t needed any more
for datasetName in localDataGroup.keys():
if datasetName not in localDatasetIds:
del localDataGroup[datasetName]
if wroteInternalData:
// Force the operator to setupOutputs() again so it gets data from the project, not external files
// TODO: This will cause a slew of "dirty" operators for any inputs we saved.
// Is that a problem?
infoCopy = copy.copy(self.mainOperator.Dataset[0].value)
self.mainOperator.Dataset[0].setValue(infoCopy)
self._dirty = False
def _deserializeFromHdf5(self, topGroup, groupVersion, hdf5File, projectFilePath):
with Tracer(traceLogger):
// The "working directory" for the purpose of constructing absolute
// paths from relative paths is the project file"s directory.
In pattern: SUPERPATTERN
Frequency: 4
Non-data size: 7
Instances
Project Name: ilastik/ilastik
Commit Name: f921a41292d13178a6eca168b8bd261f190b76b4
Time: 2012-06-26
Author: bergs@janelia.hhmi.org
File Name: ilastik-shell/applets/dataSelection/dataSelectionSerializer.py
Class Name: DataSelectionSerializer
Method Name: _serializeToHdf5
Project Name: ilastik/ilastik
Commit Name: be81d75f31d318e087760bf9cb5b80a1540f3ddd
Time: 2012-06-25
Author: bergs@janelia.hhmi.org
File Name: ilastik-shell/applets/dataSelection/dataSelectionGui.py
Class Name: DataSelectionGui
Method Name: addFileNames
Project Name: ilastik/ilastik
Commit Name: f921a41292d13178a6eca168b8bd261f190b76b4
Time: 2012-06-26
Author: bergs@janelia.hhmi.org
File Name: ilastik-shell/applets/pixelClassification/pixelClassificationSerializer.py
Class Name: PixelClassificationSerializer
Method Name: _serializePredictions
Project Name: ilastik/ilastik
Commit Name: c99c7e6af33e4e9b6b5f4564b042cafc5a440b1a
Time: 2012-06-25
Author: bergs@janelia.hhmi.org
File Name: lazyflow/operators/obsolete/classifierOperators.py
Class Name: OpTrainRandomForestBlocked
Method Name: execute
